Dilation of VOIs
Using MRIcron to investigate perfusion and diffusion values in regions
around lesions at MUSC

This page briefly describes the practical steps required to analyze MRI
diffusion and perfusion data, using
FSL,
Java Image, and
MRIcron to produce diffusion and
perfusion values from rings of dilated VOIs around a brain lesion, specifically
for the brain tumor patient study in Radiology & Radiological Sciences at
MUSC.
1. Retrieve Patients from PACS using OsiriX
-
Use the dual-monitor Mac. (Leo Bonilha will know its location if it gets
moved, or if the username and password are required.)
- Start
OsiriX

-
Click Query.
- In the new window, select the Patient ID tab. Enter the patient's
Medical Record Number (MRN).
-
In the list of the patient's appointments, select the appropriate scan date and
ensure the modality for that appointment includes MR.
-
Click Retrieve. This takes less than one minute per patient.
-
Repeat searching and retrieving all patients. Several Retrieves can be run in
parallel.
2. Export DICOM from OsiriX
-
Back in the OsiriX main window, highlight all patients for export (hold down the
Apple key while selecting multiple patients).
-
Click Export.
-
Browse to the required destination folder on the Mac's System HD (e.g.,
/morgan ).
-
Create a new folder (using the button lower left) with a unique name that does
not contain any spaces (e.g., tumor_todays_date ).
-
Click Choose. Wait for the DICOM export to finish.
3. Transfer DICOM to the CAIR hornet server
4. Connect to the CAIR hornet server using VNC
5. Process perfusion data using JIM
-
Change into the patient subdirectory on hornet. Identify the file containing the
perfusion raw data. If the DICOM files were
converted using
dcm2nii
then the filename should indicate the name of the acquisition protocol used. If
dtoa was used for conversion, type
showana *.hdr
| grep desc to list the acquisition protocol used for each image file.
-
The raw perfusion data file will contain a name similar to ep2d_perf
if acquired on a Siemens MR scanner, or PERF/PRESTO if acquired on a
Philips MR scanner.
-
However, more than one image file may contain this identifier,
as this label may also be attached to images produced by the scanner's on-line
perfusion processing package. If this is the case, each image file containing
the perfusion label should be inspected. The raw perfusion image file will
contain multiple dynamic volume measurements, whereas the on-line perfusion
processed results will normally only contain one volume. To determine the
number of dynamic volumes, either type
showana image_filename and
examine the line starting 'images per volume', or use graphical software such as
MRIcro to display the number of volumes.
-
Once the perfusion raw data image file has been indentified, display related
information required by the perfusion processing software by typing
showana image_filename In particular, note the TE from the
'description' line, the 'number of volumes', and the 'volume duration'.
-
Start the Java Image software by typing
jim &
-
The first time you run Jim, I suggest modifying some of the preferences as
follows. Select Toolkits / Preferences. Under the Misc tab,
set
- I work most often with: Analyze images
- I like my NIFTI images in: One file .nii
- I like my NIFTI images: Compressed
- Also select a more relevant startup directory.
-
Click Save Settings on this tab, then select the
Interoperability tab.
- Tick the Create new Analyze 7.5 images only with non-flipped
orientations box
-
then click Save Settings and Done.
5.3 Perfusion Analysis
-
From the main menu, select Toolkits / Brain Perfusion. In the new
Perfusion Analysis
window, accept the default parameter values except as discussed below.
- Input configuration: Select Single input image. Click on the
folder icon then browse and select the perfusion raw data file that you
identified. above. File browsing with JIM in general can take a bit of getting
used to, in particular, it appears best to always browse for a file graphically
(rather than typing its name in manually), and when double clicking to select
a sub-directory, it's more successfully to double click on the folder icon to the left of the folder name rather than on the folder name itself.
- Input configuration: The Number of time points in image is the
number of volumes displayed by
showana image_filename
above.
- Input configuration: The Time between images can usually be
determined from the volume duration displayed by
showana
image_filename above. It should be in the range of about 0.5 - 4
seconds. Depending on the source DICOM file, the volume duration
displayed by showana may be correct, or it may be the time for the
entire acquisition in which case you will need to divide it by the
number of volumes first. Also remember to convert from ms to s.
- Input configuration: Leave the Number of steady state images as 1
as for the protocols at MUSC these will alreayd have been discarded as
'dummies' on the scanner. Select Register images to perform motion
correction. Optionally select Apply smoothing filter (but once you
have decided whether to use it or not, be consistent for all subjects). Rather
than use Brain Finder, you can define a brain mask by applying FSL's
BET to the first volume of the raw perfusion data after the JIM perfusion
analysis has finished.
- Threshold: Typically a value of 80 should be okay.
- Quantification parameters: For the Scan TE enter the TE value from
the description line displayed by
showana above. Leave
the other parameters in this section at their default values.
- Arterial Input Function: The delay between start of the scan and the
first arrival of the contrast agent in the brain can vary between patients
depending on their heart rate, age, and the relative time that the injection
was actually initiated. The parameters listed here appear to work for the
current perfusion protocol at MUSC, and one would hope that the MRI techs
always time the perfusion scans consistently. However, this may not always be
the case, not least due to new MR scanners being installed, software upgrades,
or the decision to modify the perfusion protocol. During the perfusion analysis,
below, the AIF graph is displayed on the screen containing dots to indicate
at what point the analysis started to look for contrast. If this is right at the
start of the rising curve, you should probably reduce the Scan number at
contrast arrival by 2 and re-run the analysis. Likewise, if the dot on the
graph is well before the curve starts to rise, the initial pre-contrast baseline
interval may be too short resulting in a poor estimate of the baseline value
which will affect the perfusion calculation, and so the Scan number at
contrast arrival should be increased and the analysis re-run.
Example of varying Scan Number At Contrast Arrival parameter
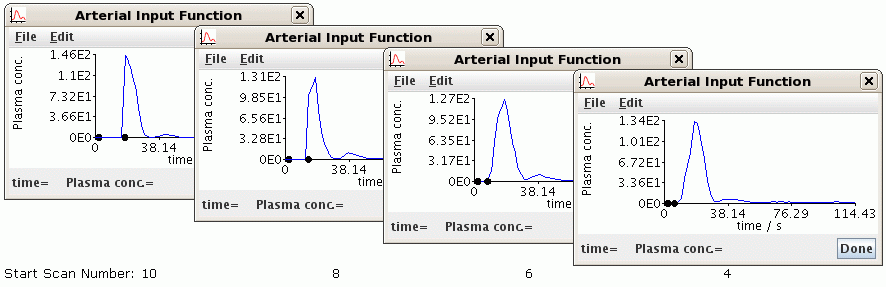
Try a value of 8 for Scan number at contrast arrival and a value of
one less than the number of dynamic volumes, for the Analysis end scan
number.
When you become more familiar using JIM, there is also an option to plot the
timecourse of individual pixels, or ROIs, using the
Dynamic Analysis - Roaming
Intensities tool.
- AIF selection: Automatic.
- Output folder: Select this graphically, to be the same as the folder
containing the input raw perfusion image files. Click Load to
accept the folder selection.
- Output base name: Type this in manually. I suggest appending "_perf" to the
input filename, so if the input filename is
set09.hdr the use an
Output base name of set09_perf
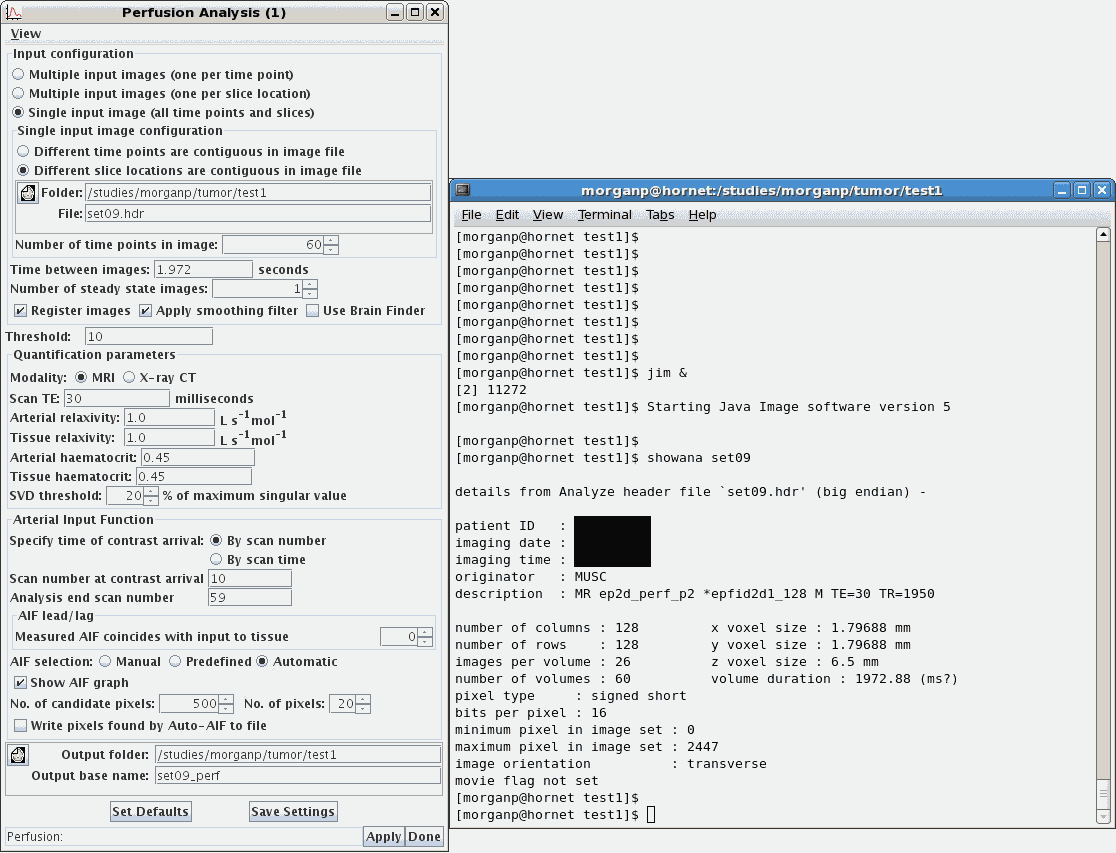
- Click Apply to start the analysis.
-
Load one of the perfusion images (e.g., the CBV map) into an image
viewing program to check they look reasonable. Do not use JIM for this, but use
MRIcron,
MRIcro,
fslview, or
ibx, all of which are available on the hornet server.
Check that the images appear reasonable and are the right way up. If the images
are upside down, check your preference settings as described in
section 5.2, above, and then re-run the perfusion analysis.
-
Create a brain mask. For a later stage of analysis, below, this mask needs to be
in MRIcron VOI format. Create this mask as follows.
-
Start MRIcron by typing
mricron &
-
Select File / Open and load in the raw perfusion scans. As this is a
dynamic multi-volume acquisition, you will be prompted for which volume number
to load. Select the first volume.
-
Select Draw / Advanced / Brain Extraction. Reduce the default 0.5
factor to 0.2 and click Go.
- If the resulting images look reasonable, then close the Brain Extraction
window and select Draw / Advanced / Brain Mask. This will prompt to
save the brain mask in VOI format. Choose a filename with the same base name as the perfusion acquisition, e.g., if the perfusion raw data were in a
file named set09 then name the VOI file something like
set09_brain_mask
-
Usually the Volume Of Interest will be drawn on the FLAIR image. Indentify which
file contains the FLAIR using a similar process as in
section 5.1, above, i.e., by typing
showana
*.hdr | grep desc
-
FLAIR scans acquired on a Siemens scanner may contain the name FLAIR or
tir2d in the decription field. Acquisitions on a Philips scanner are
usually labeled FLAIR.
-
Start MRIcron by typing
mricron &
- Check the options in the Help / Preferences page. Ensure that the
Show drawing menus and tools option is selected. Also note the
Reorient images when loading option. This should be unselected before
loading the FLAIR images for drawing the VOIs. However, selecting it and
reloading the FLAIR scans may make visual assessment of the lesion and final
VOI easier.
- Select File / Open to load the FLAIR image file.
-
Scroll through the slices and identify the lesion. Draw round the lesion, as
instructed by the neuroradiologist, slice by slice.
-
Draw by first clicking on the Autoclose pen button
 then click and hold while drawing with the mouse. After completing the region
on each slice, fill the region by either right clicking in it, or by using the
Fill tool button.
then click and hold while drawing with the mouse. After completing the region
on each slice, fill the region by either right clicking in it, or by using the
Fill tool button.
- Repeat for all slices. Save by selecting Draw / Save VOI...
-
(To view the VOI in interpolated 3D, select the Reorient images when
loading option, see above, then reload the FLAIR scans, then select
Draw / Open to reload the VOI file. Scroll through the orthogonal
slices by clicking the crosshair.)
-
Start MRIcron and load the processed perfusion map (e.g., CBV or CBF).
-
Load the VOI drawn on the FLAIR, above, by selecting Draw / Open VOI.
Especially if using a combination of NIFTI images, check that a left-right flip
has not occurred, and that the VOI overlays as expected. Note, this stage is
just a visual check; you will re-select these images again during the dilation
process below.
-
Select Draw / Advanced / Dilate VOIs
-
The next prompt asks for the number of boundaries for the dilations around the
surface of the VOI. So, for example, to obtain four adjacent dilated bands
around the core VOI, enter 5 edges.
-
Enter the distance of the edges of the dilations away from the VOI surface.
Entering 0, 5, 10, 15, 20 will result in four dilated bands around the core VOI,
where band 1 ranges from 0-4 mm, band 2 from 5-9 mm, band 3 from 10-14 mm, and
band 4 from 15-19 mm. Dilations occur in 3D.
-
You then have the option to mask the dilated VOIs with an additional mask. This
is useful to prevent the dilations extending outside the brain, or to
constrain the dilations to a particular hemisphere, or to just white matter. In
the case, select Yes and choose the brain mask created above in
section 5.4. To select the VOI file containing the brain mask
created above, first change the File of type: pull down menu to
Volume of interest (*.voi).
-
In the next Select VOI browser window, reselect the VOI file that
outlines the core lesion.
-
The next Select PERF image browser window requires the filename of the
post-processed perfusion image to analyse (CBV, CBF, etc.).
-
The dilated VOIs are then generated, and saved. The next Select VOI
browser
window is prompting to repeat the analysis with a different core lesion VOI.
Click Cancel, and a Descriptive Statistics window will open containing
the results from each dilated band around the core lesion. It may be easier to
save these data (File / Save) and view the results in another editor or
spreadsheet. the values in this file are the values from whichever perfusion
parametr file was selected above, e.g., CBV, CBF, etc.
8. Non-perfusion data
-
While the instructions on this page relate to analysis of perfusion images, the
same technique can be applied to other parameter maps, such as diffusion maps
of ADC (or MD). The perfusion maps will need to be generated, but then the
dialation stage is the same as in section 7, above, except
that instead of selecting perfusion parameter map of CBV or CBF, instead select
the diffusion ADC image.
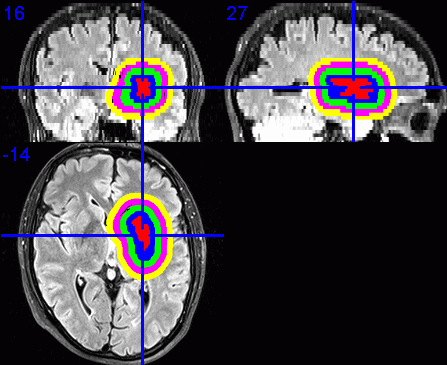
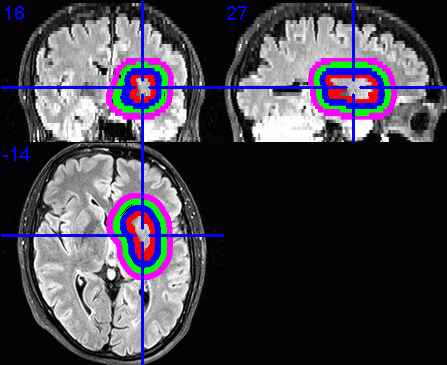
9. Visualisation of dilated VOIs
-
To visualise the dilated VOIs using MRIcron, I suggest first enabling the
Reorient images when loading setting, as described in the fourth
point in section 6 above.
-
Load the required base images, e.g., the FLAIR images.
-
Select Overlay / Add. Change the Files of type: to VOIs and
choose the core lesion VOI file.
-
Repeat the step above, expect each time select the VOI file for the next
dilated band. The VOI filenames start with a 1, 2, 3, ... depending on the
level of dilation, followed by the name of the core VOI. Each VOI should be
displayed in a different colour.
-
Click on the centre of the lesion to obtain the required view of the VOIs in
the three orthogonal planes.
- Select File / Save as Bitmap to save this view as an image that
could be inserted into a document or presentation.